Our Simulations
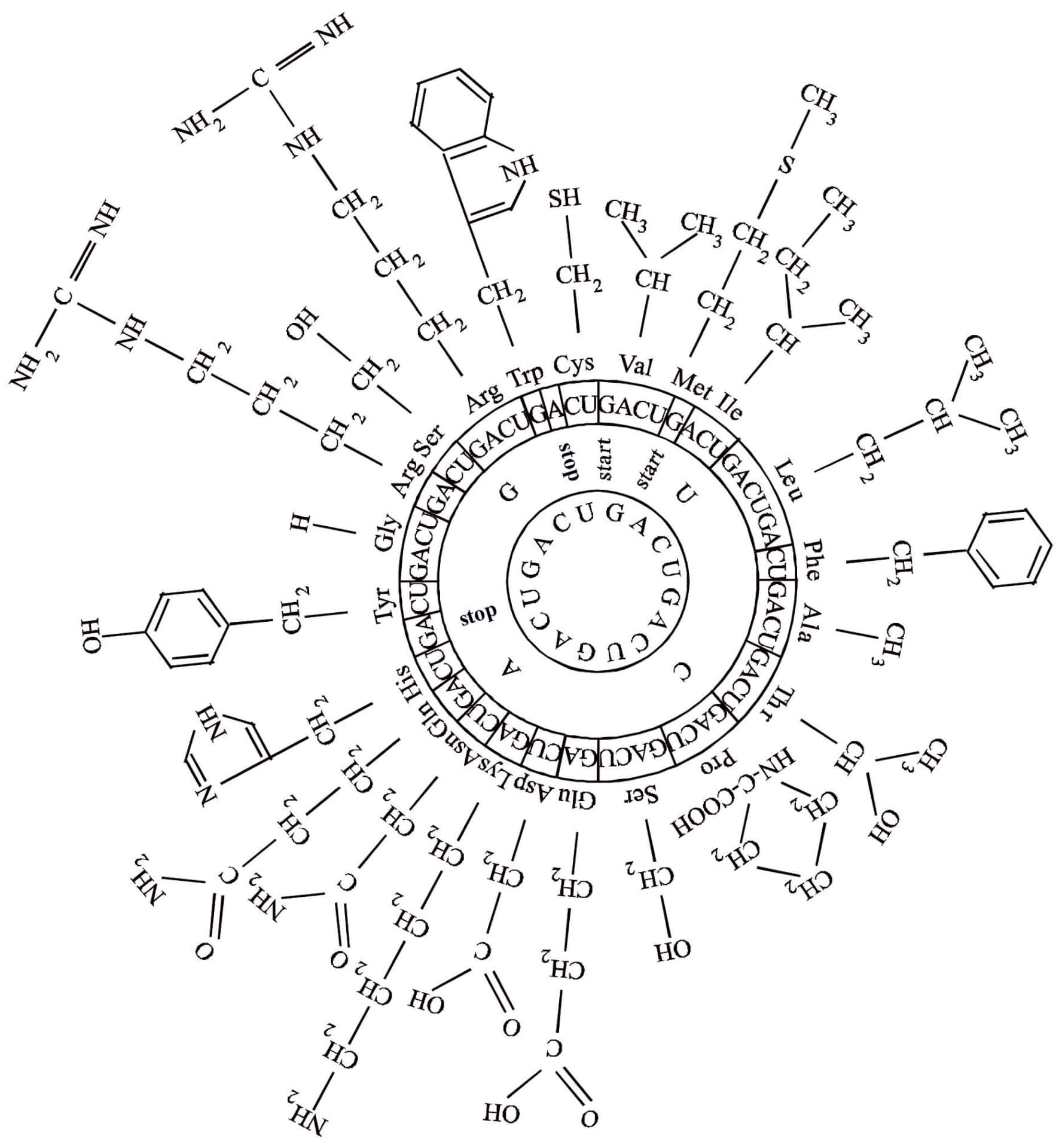
Click on a particular toggle on the right, e.g., A: #class = 4, to dropdown a list of results of my experiments. You can display them or download their ‘zip‘ files to inspect them, e.g., in Notepad++ with line numbers.

Results
Input parameters are loaded from settings.txt every time my program tRNALab is launched. They are also listed at the end of each output file. #class means the prescribed number of classificatory groups. All input parameters in capitals are Boolean. (The last section of settings.txt, entitled by a-check-of-included-positions, is updated after each start of tRNALab in dependence on the previous parameters.)
Two compilations of tRNA genes were examined for control: 1) the Leipzig database and 2) the database by USCS.
Any presence of classifications 1) XMITNKRF, 2) HQE, 3) DAV, 4) LSYCUW, 5) G, and 6) P (Eq. 2) for #class = 6 or 1) XMITNKRF, 2) QE, 3) DAV, 4) HLSU, 5) YWC, 6) G, and 7) P (Eq. 4) for #class = 7 is indicated by + on this page and in output listings.
Any presence of classifications 1) XMITNKR, 2) HQE, 3) DAV, 4) FLSYCUW, 5) G, and 6) P (Eq. 3a) for #class = 6 or 1) XMITNKR, 2) HQE, 3) DAV, 4) LSU, 5) FYCW, 6) G, and 7) P (Eq. 5a) for #class = 7 is indicated by ++ on this page and in output listings.
Functions: f0 = FindLUCA, f1 = FindScores, f2 = FindNonStartAAs, f3 = FindNonGlobAAs, f4 = GlobOptimizeGroups, and f5 = CompleteTestData.
You can download a tree of all results packed in zip.
The Leipzig database of tRNA genes:
Non-standard assignments = 0 (false) (NS0):
There are 37 tRNA genes with non-standard assignments in the Leipzig database, e.g., 14 tRNA genes of Ile are assigned to a codon AUG. These 37 tRNA genes are ignored in the following experiments. Data = abe_L_dataD, zip. Here, ‘a‘ represents archaea, ‘b‘ – bacteria, ‘e‘ – eukaryotes, ‘L‘ – the Leipzig database, and ‘D‘ – tRNA genes.
- f4, sites = all (settings, zip, abe_L_classifications, zip, abe_L_settings_with_comments.txt, abe_L_chronologies, abe_L_p_simChart, abe_L_ch_p_simChart, job.out, zip)
- f4, sites = all (settings, zip, abe_L_classifications, zip, abe_L_chronologies, abe_L_p_simChart, abe_L_ch_p_simChart, job.out, zip)
- f4, sites = all (settings, zip, abe_L_classifications, zip, abe_L_chronologies, abe_L_p_simChart, abe_L_ch_p_simChart, job.out, zip) Eq. 2 of the manuscript +
- f3, sites = all (settings, zip, abe_L_nonGlobAAs, zip, job.out, zip) ⇒ ¬Glob = CU is set as an input parameter of (C.6) ++
- f3, sites = all, MX_MOVE_TOGETHER = 1 (settings, zip, abe_L_nonGlobAAs, zip, job.out, zip) ⇒ ¬Glob = CU ++
- f3, sites = G3 (settings, zip, abe_L_nonGlobAAs, zip, job.out, zip)
- f3, sites = G3, MX_MOVE_TOGETHER = 1 (settings, zip, abe_L_nonGlobAAs, zip, job.out, zip)
- f4, sites = all, ¬Glob = CU (settings, zip, abe_L_classifications, zip, abe_L_chronologies, abe_L_p_simChart, abe_L_ch_p_simChart, job.out, zip) Eq. 3 ++
- f2, sites = all (settings, zip, abe_L_nonStartAAs, zip) 7 ✕ +
- f2, sites = all, MX_MOVE_TOGETHER = 1 (settings, zip, abe_L_nonStartAAs, zip) 9 ✕ +
- f4, sites = all (settings, zip, abe_L_classifications, zip, abe_L_chronologies, abe_L_p_simChart, abe_L_ch_p_simChart, job.out, zip)
- f3, sites = all (settings, zip, abe_L_nonGlobAAs, zip, job.out, zip) ⇒ ¬Glob = CUH is set as an input parameter of (D.6) + !!!!!!!!!!!!!!!!!!!!!!!!!!!!!!
- f3, sites = all, MX_MOVE_TOGETHER = 1 (settings, zip, abe_L_nonGlobAAs, zip, job.out, zip) ⇒ ¬Glob = CUH ++
- f3, sites = G3 (settings, zip, abe_L_nonGlobAAs, zip, job.out, zip)
- f3, sites = G3, MX_MOVE_TOGETHER = 1 (settings, zip, abe_L_nonGlobAAs, zip, job.out, zip) +
- f4, sites = all, ¬Glob = CUH (settings, zip, abe_L_classifications, zip, abe_L_chronologies, abe_L_p_simChart, abe_L_ch_p_simChart, job.out, zip) Eq. 5 ++
- f4, sites = all (settings, zip, abe_L_classifications, zip, abe_L_chronologies, abe_L_p_simChart, abe_L_ch_p_simChart, job.out, zip)
- f4, sites = all (settings, zip, abe_L_classifications, zip, abe_L_chronologies, abe_L_p_simChart, abe_L_ch_p_simChart, job.out, zip)
- f4, sites = all (settings, zip, abe_L_classifications, zip, abe_L_chronologies, abe_L_p_simChart, abe_L_ch_p_simChart, job.out, zip)
Non-standard assignments = 1 (true) (NS1):
There are 37 tRNA genes with non-standard assignments in the Leipzig database, e.g., 14 tRNA genes of Ile are assigned to a codon AUG. They are treated as if they were assigned to a codon AUU in the following experiments. The rest of the non-standard assignments are treated analogically. Data = abe_L_dataD, zip.
- f4, sites = all (settings, zip, abe_L_classifications, zip, abe_L_chronologies, abe_L_p_simChart, abe_L_ch_p_simChart, job.out, zip)
- f4, sites = all (settings, zip, abe_L_classifications, zip, abe_L_chronologies, abe_L_p_simChart, abe_L_ch_p_simChart, job.out, zip)
- f0, sites = all (settings, zip, abe_L_FindLUCA, zip)
- f4, sites = all (settings, zip, abe_L_classifications, zip, abe_L_chronologies, abe_L_p_simChart, abe_L_ch_p_simChart, job.out, zip) Eq. 2 +
- f3, sites = all (settings, zip, abe_L_nonGlobAAs, zip, job.out, zip) ⇒ ¬Glob = CU is set as an input parameter of (J.7) ++
- f3, sites = all, MX_MOVE_TOGETHER = 1 (settings, zip, abe_L_nonGlobAAs, zip, job.out, zip) ⇒ ¬Glob = CU ++
- f3, sites = G3 (settings, zip, abe_L_nonGlobAAs, zip, job.out, zip) +
- f3, sites = G3, MX_MOVE_TOGETHER = 1 (settings, zip, abe_L_nonGlobAAs, zip, job.out, zip) +
- f4, sites = all, ¬Glob = CU (settings, zip, abe_L_classifications, zip, abe_L_chronologies, abe_L_p_simChart, abe_L_ch_p_simChart, job.out, zip) Eq. 3-4 ++
- f2, sites = all (settings, zip, abe_L_nonStartAAs, zip) 7 ✕ +
- f2, sites = all, MX_MOVE_TOGETHER = 1 (settings, zip, abe_L_nonStartAAs, zip) 10 ✕ +
- f4, sites = all (settings, zip, abe_L_classifications, zip, abe_L_chronologies, abe_L_p_simChart, abe_L_ch_p_simChart, job.out, zip)
- f3, sites = all (settings, zip, abe_L_nonGlobAAs, zip, job.out, zip) ⇒ ¬Glob = CUH is set as an input parameter of (K.6) ++
- f3, sites = all, MX_MOVE_TOGETHER = 1 (settings, zip, abe_L_nonGlobAAs, zip, job.out, zip) ⇒ ¬Glob = CUH ++
- f3, sites = G3 (settings, zip, abe_L_nonGlobAAs, zip, job.out, zip) +
- f3, sites = G3, MX_MOVE_TOGETHER = 1 (settings, zip, abe_L_nonGlobAAs, zip, job.out, zip) +
- f4, sites = all, ¬Glob = CUH (settings, zip, abe_L_classifications, zip, abe_L_chronologies, abe_L_p_simChart, abe_L_ch_p_simChart, job.out, zip) Eq. 5, Extended Fig.1, Fig. 2 ++
- f4, sites = all (settings, zip, abe_L_classifications, zip, abe_L_chronologies, abe_L_p_simChart, abe_L_ch_p_simChart, job.out, zip)
- f4, sites = all (settings, zip, abe_L_classifications, zip, abe_L_chronologies, abe_L_p_simChart, abe_L_ch_p_simChart, job.out, zip)
- f4, sites = all (settings, zip, abe_L_classifications, zip, abe_L_chronologies, abe_L_p_simChart, abe_L_ch_p_simChart, job.out, zip)
The database of tRNA genes by UCSC:
Please, keep in mind that this database contains no tRNA genes of rare selenocysteine (Sec ≡ U).
Non-standard assignments = 0 (false) (NS0):
Data = abe_C_dataD, zip.
- f4, sites = G3, score ≧ 67, MX_MOVE_TOGETHER = 1 (settings, zip, abe_C_classifications, zip, abe_C_chronologies, abe_C_p_simChart, abe_C_ch_p_simChart, job.out, zip)
- f4, sites = G3, score ≧ 67, MX_MOVE_TOGETHER = 1 (settings, zip, abe_C_classifications, zip, abe_C_chronologies, abe_C_p_simChart, abe_C_ch_p_simChart, job.out, zip)
- f0, sites = all, score ≧ 67, MX_MOVE_TOGETHER = 1 (settings, zip, abe_C_FindLUCA, zip)
- f1, sites = all, MX_MOVE_TOGETHER = 1 (settings, zip, abe_C_FindScores, zip) ⇒ score ≧ 67 is set as an input parameter of (O.1, P.1, Q.1, Q.3, Q.4, Q.8-U.1) ++
- f1, sites = G3, MX_MOVE_TOGETHER = 1 (settings, zip, abe_C_FindScores, zip) ⇒ score ≧ 67.5-69.5 ++
- f4, sites = G3, score ≧ 67, MX_MOVE_TOGETHER = 1 (settings, zip, abe_C_classifications, zip, abe_C_chronologies, abe_C_p_simChart, abe_C_ch_p_simChart, job.out, zip)
- f3, sites = all, score ≧ 67 (settings, zip, abe_C_nonGlobAAs, zip, job.out, zip)
- f3, sites = all, score ≧ 67, MX_MOVE_TOGETHER = 1 (settings, zip, abe_C_nonGlobAAs, zip, job.out, zip) ⇒ ¬Glob = ∅ ++
- f4, sites =all, score ≧ 67, MX_MOVE_TOGETHER = 1, ¬Glob = ∅ (settings, zip, abe_C_classifications, zip, abe_C_chronologies, abe_C_p_simChart, abe_C_ch_p_simChart, job.out, zip) ++
- f3, sites = G3, score ≧ 67 (settings, zip, abe_C_nonGlobAAs, zip, job.out, zip) ⇒ ¬Glob = Q ++
- f3, sites = G3, score ≧ 67, MX_MOVE_TOGETHER = 1 (settings, zip, abe_C_nonGlobAAs, zip, job.out, zip) ⇒ ¬Glob = Q is set as an input parameter of (Q.10) ++
- f4, sites = G3, score ≧ 67, MX_MOVE_TOGETHER = 1, ¬Glob = Q (settings, zip, abe_C_classifications, zip, abe_C_chronologies, abe_C_p_simChart, abe_C_ch_p_simChart, job.out, zip) Eq. 6 ++
- f4: sites = G3, score ≧ 67, MX_MOVE_TOGETHER = 1 (settings, zip, abe_C_classifications, zip, abe_C_chronologies, abe_C_p_simChart, abe_C_ch_p_simChart, job.out, zip)
- f3: sites = all, score ≧ 67 (settings, zip, abe_C_nonGlobAAs, zip, job.out, zip)
- f3: sites = all, score ≧ 67, MX_MOVE_TOGETHER = 1 (settings, zip, abe_C_nonGlobAAs, zip, job.out, zip)
- f3: sites = G3, score ≧ 67 (settings, zip, abe_C_nonGlobAAs, zip, job.out, zip)
- f3: sites = G3, score ≧ 67, MX_MOVE_TOGETHER = 1 (settings, zip, abe_C_nonGlobAAs, zip, job.out, zip) ⇒ ¬Glob = CQE is set as an input parameter of (R.6) ++
- f4, sites = G3, score ≧ 67, MX_MOVE_TOGETHER = 1, ¬Glob = CQE (settings, zip, abe_C_classifications, zip, abe_C_chronologies, abe_C_p_simChart, abe_C_ch_p_simChart, job.out, zip) Eq. 7 ++
- f4: sites = G3, score ≧ 67, MX_MOVE_TOGETHER = 1 (settings, zip, abe_C_classifications, zip, abe_C_chronologies, abe_C_p_simChart, abe_C_ch_p_simChart, job.out, zip)
- f4: sites = G3, score ≧ 67, MX_MOVE_TOGETHER = 1 (settings, zip, abe_C_classifications, zip, abe_C_chronologies, abe_C_p_simChart, abe_C_ch_p_simChart, job.out, zip)
- f4: sites = G3, score ≧ 67, MX_MOVE_TOGETHER = 1 (settings, zip, abe_C_classifications, zip, abe_C_chronologies, abe_C_p_simChart, abe_C_ch_p_simChart, job.out, zip)
Although there are 1271 tRNA genes with non-standard assignments in the database by UCSC provided that score ≧ 67, they are all assigned to Ile and do not significantly change results of our simulations. Therefore, a subsection “Non-standard assignments = 1 (true) (NS1)” is missing here, unlike the list of results mentioned above for the Leipzig database.
Functions: f0 = FindLUCA, f1 = FindScores, f2 = FindNonStartAAs, f3 = FindNonGlobAAs, f4 = GlobOptimizeGroups, and f5 = CompleteTestData.